Dec. 4, 2012, 7:11 a.m. by dewshick
Topics: Heredity, Phylogeny, Probability
Lying in Wait
Single gene disorders can be encoded by either dominant or recessive alleles. In the latter case, the affected person usually has two healthy carrier parents, who were usually unaware that their child could inherit a deadly or debilitating genetic condition from them.
We know from Mendel's first law that any offspring of two heterozygous carriers has a 25% chance of inheriting a recessive disorder. Knowing your own genotype is therefore important when deciding to have children, and genetic screening will prove vital for preventive medicine in the coming years.
In this problem, we will consider an exercise in which we determine the probability of an organism exhibiting each possible genotype for a factor knowing only the genotypes of the organism's ancestors.
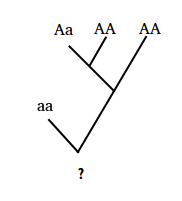
A rooted binary tree can be used to model the pedigree of an individual. In this case, rather than time progressing from the root to the leaves, the tree is viewed upside down with time progressing from an individual's ancestors (at the leaves) to the individual (at the root).
An example of a pedigree for a single factor in which only the genotypes of ancestors are given is shown in Figure 1.
Given: A rooted binary tree
Return: Three numbers between 0 and 1, corresponding to the respective probabilities that
the individual at the root of
((((Aa,aa),(Aa,Aa)),((aa,aa),(aa,AA))),Aa);
0.156 0.5 0.344