April 17, 2013, 6 p.m. by Rosalind Team
Topics: Combinatorics, Phylogeny
Seeing the Forest
In “Counting Unrooted Binary Trees”, we found a way to count the number of unrooted binary trees representing phylogenies on
$n$ taxa. Our observation was that two such trees are considered distinct when they do not share the same collection of splits.Counting all these trees is one task, but actually understanding how to write them out in a list (i.e., enumerating them) is another, which will be the focus of this problem.
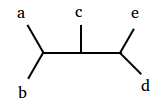
Recall the definition of Newick format from “Distances in Trees” as a way of encoding trees. See Figure 1 for an example of Newick format applied to an unrooted binary tree whose five leaves are labeled (note that the same tree can have multiple Newick representations).
Given: A collection of species names representing
Return: A list containing all unrooted binary trees whose leaves are these
dog cat mouse elephant
(((mouse,cat),elephant))dog; (((elephant,mouse),cat))dog; (((elephant,cat),mouse))dog;